Detection of donor-derived cell-free DNA: Non-invasive prediction of silent antibody-mediated rejection in donor-specific antibody-positive renal allograft recipients
Markus Wahrmann1, Thierry Viard2, Amanda Tillgren2, Natali Gulbahce2, Deanna Lee2, Nicolas Kozakowski3, Heinz Regele3, Helmuth Haslacher4, Farsad Eskandary1, Jonathan McHugh2, Hal Gibson2, Marica Grskovic2, Georg A. Böhmig1.
1Department of Medicine III, Medical University of Vienna, Vienna, Austria; 2CareDx Inc., Brisbane, San Francisco, CA, United States; 3Department of Pathology, Medical University of Vienna, Vienna, Austria; 4Department of Laboratory Medicine, Medical University of Vienna, Vienna, Austria
Background: The detection of donor-specific antibodies (DSA) has limited value as a diagnostic marker of late antibody-mediated rejection (ABMR), and the presence of circulating DSA does not necessarily indicate an ongoing rejection process. Here, we investigated whether and to which extent analysis of donor-derived cell-free DNA (dd-cfDNA) helps improve non-invasive monitoring of silent ABMR.
Methods: The study cohort consisted of 45 HLA class I and/or II DSA-positive kidney allograft recipients identified upon systematic cross-sectional alloantibody/ABMR screening (protocol biopsies in case of a positive DSA result). Twenty-five of these patients were diagnosed with ABMR (Banff 2017), and 20 recipients had no rejection. Cell-free DNA was extracted from 0.5 mL biobanked EDTA-plasma using Qiagen’s QIAamp Circulating Nucleic Acid Kit, followed by a double AMPure clean-up step to remove contaminating cell-derived DNA. Cell-free DNA was then analyzed by CareDx AlloSeq cfDNA assay sequencing.
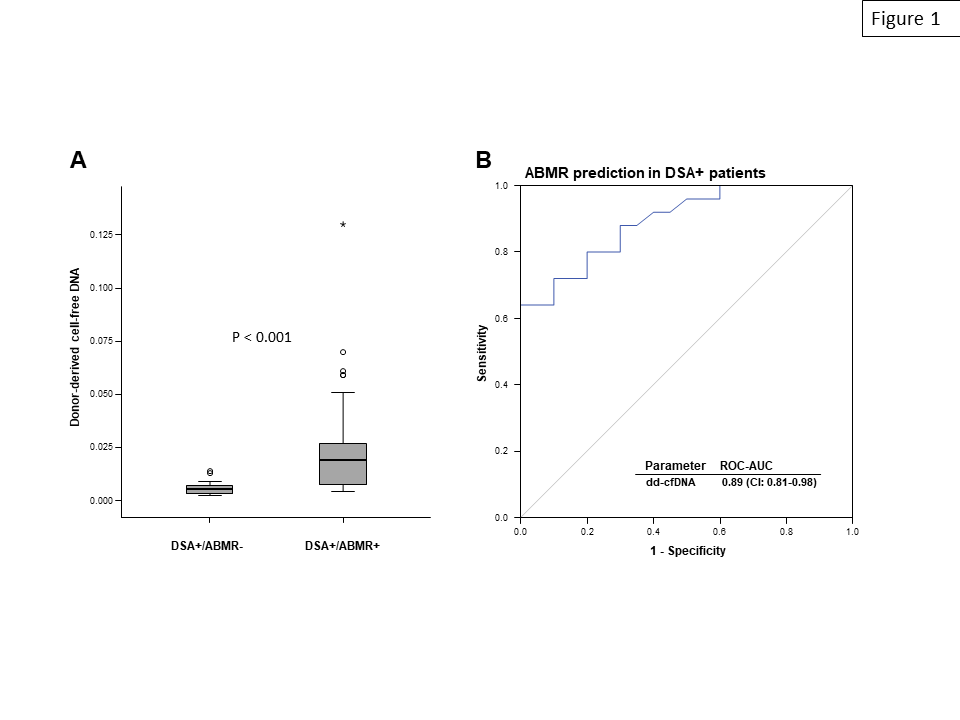
Results: Overall, DSA-positive study subjects showed a significantly higher median percentage of dd-cfDNA than a control group of DSA-negative transplant recipients [0.78 (IQR: 0.52-1.95) versus 0.33 (0.27-1.12), P=0.002]. DSA-positive patients diagnosed with ABMR had by far higher levels than those without rejection [%dd-cfDNA: 1.90 (0.78-3.90) versus 0.52 (0.35-0.72); P<0.001] (Figure 1A). As shown in Figure 1B, receiver operating characteristic (ROC) curve analysis for ABMR prediction revealed an area under the curve (AUC; 0.89), which exceeded AUC levels computed for the mean fluorescence intensity of immunodominant DSA (0.77). In ROC analysis, dd-cfDNA showed several points of equal highest diagnostic accuracy (0.80) including one (threshold: 1.45%) with 100% specificity and 64% sensitivity.
Conclusions: Our results support the use of dd-cfDNA monitoring, as an adjunct to DSA analysis, as a reliable non-invasive tool to uncover silent ABMR.
There are no comments yet...