
86-180 0807 0435
The prediction of kidney allograft acute rejection based on recipient peripheral blood whole transcriptome profiling and three mRNA molecular signatures
Minxue Liao1,2, Qiang Fu1,2,3, Guoli Huai1,2, Yanzhuo Liu1,2, Xiangming Quan1,2, Liang Wei1,2, Hongji Yang2,3, Gaoping Zhao1,2,3, Shaoping Deng1,2,3.
1School of Medicine, University of Electronic Science and Technology of China, Chengdu, People's Republic of China; 2Clinical Immunology Translational Medicine Key Laboratory of Sichuan Province, Sichuan Provincial People’s Hospital, University of Electronic Science and Technology of China, Chengdu, People's Republic of China; 3Organ Transplantation Center, Sichuan Provincial People's Hospital, University of Electronic Science and Techchnology of China , Chengdu, People's Republic of China
School of Medicine of University of Electronic Science and Technology of China. Clinical Immunology Translational Medicine Key Laboratory of Sichuan Province.
Introduction: This study aims to assess the ability of machine learning (ML) classifiers which based on peripheral blood transcriptome data, a timely and non-invasive approach, for the classification of patients with kidney allograft acute rejection (AR) and with stable function (STA) posttransplant.
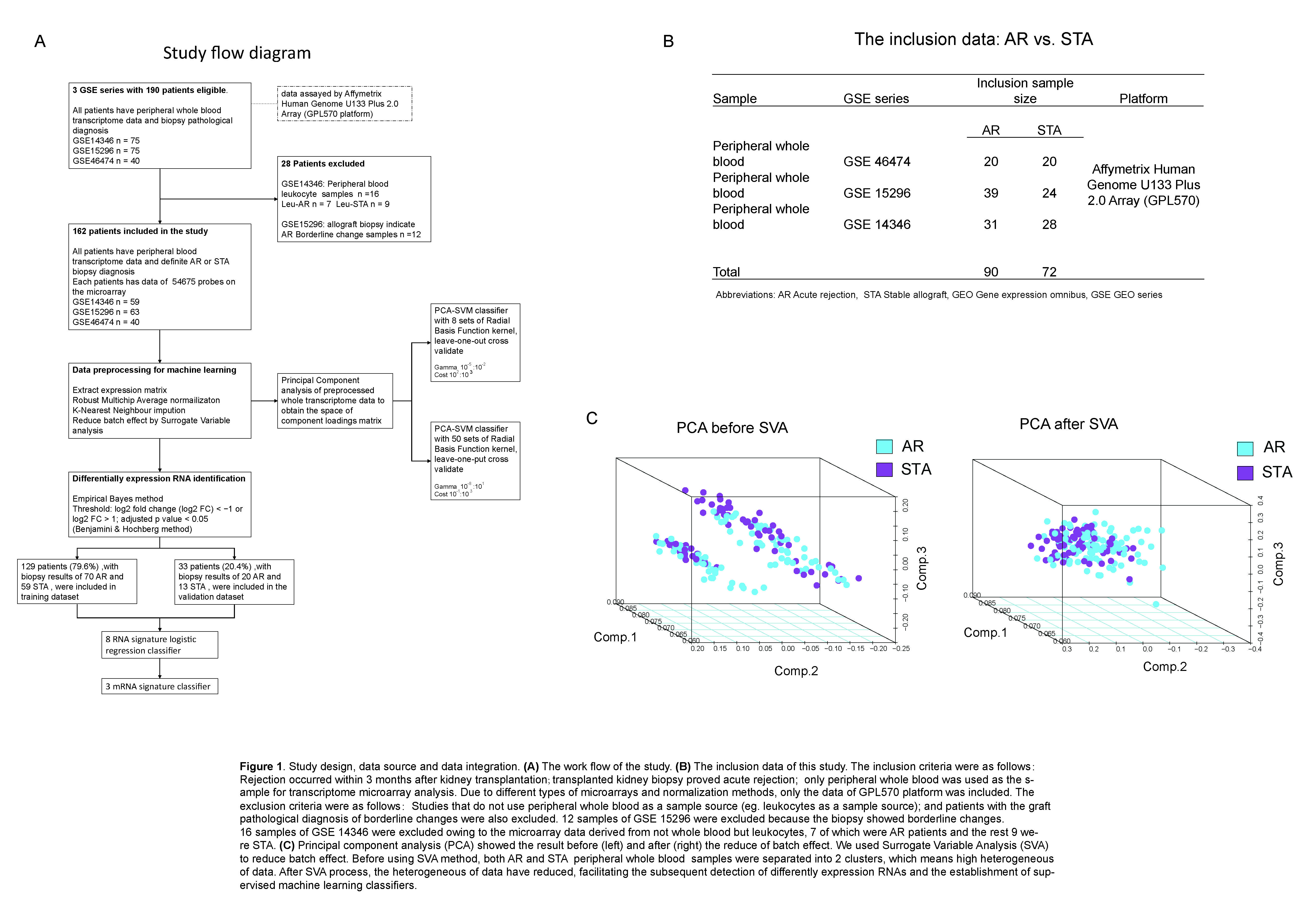
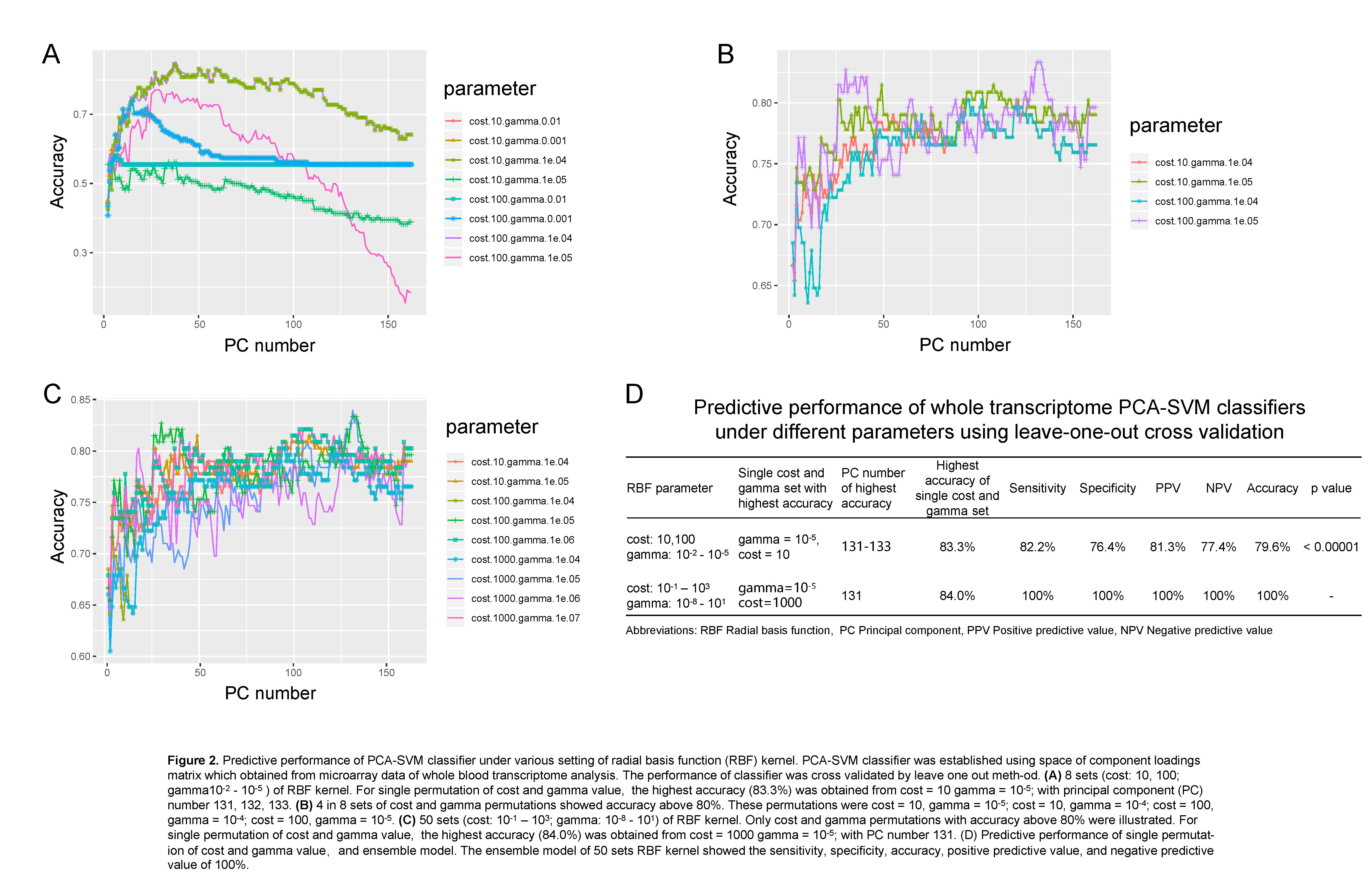
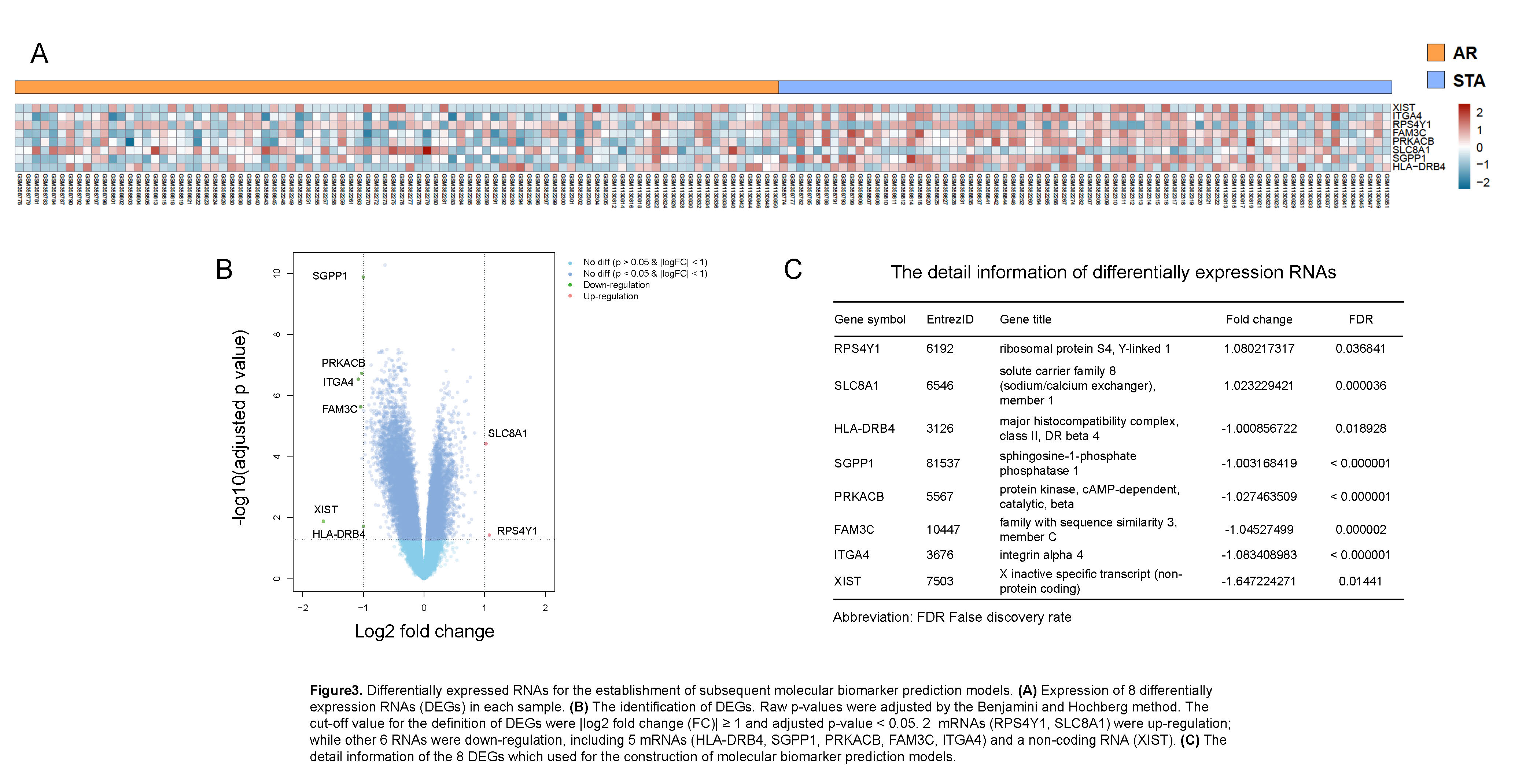
Materials and Methods: 90 AR and 72 STA patient transcriptome and pathological data updated on Gene Expression Omnibus were included. Batch effect was removed by Surrogate Variable analysis[1]. Then 3 kinds of supervised machine learning (ML) model were performed. Support Vector Machine (SVM) classifiers were constructed using the space of component loadings matrix of whole transcriptome principal component analysis (PCA), and cross validated by leave-one out method. Logistic regression classifier was based on 8 differentially expression RNAs (DEGs) identified by Empirical Bayes method. Step regression was used to assay key molecular signatures in the 8 DEGs. We evaluated the accuracy, sensitivity, and specificity of classifiers which based on whole transcriptome data, logistic regression, and step regression.
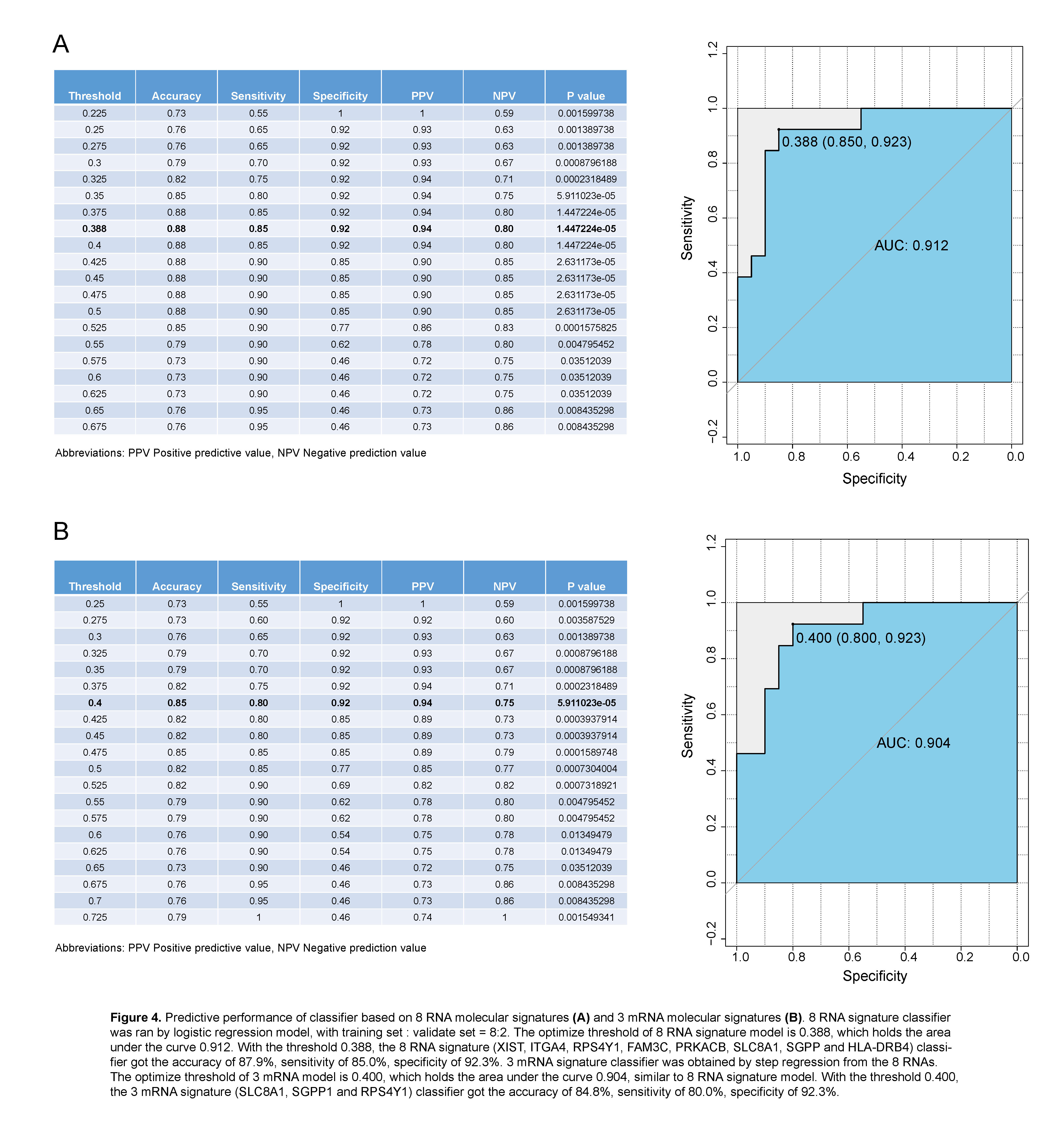
Results: We found that a classifier combined with SVM and Principal Component analysis (PCA-SVM) could better distinguish patients with AR and STA condition, with the accuracy, sensitivity and specificity of 100%. Furthermore, 8 RNA molecular signatures (XIST, ITGA4, RPS4Y1, FAM3C, PRKACB, SLC8A1, SGPP and HLA-DRB4) identified from peripheral blood could also be used to classify AR and STA patients (accuracy 87.9%, sensitivity 85.0%, specificity 92.3%, and the area under the curve (AUC) is 0.912. Step regression revealed that using 3 mRNA signatures (SLC8A1, SGPP1 and RPS4Y1) was able to detect AR patients with similar performance (accuracy 84.8%, sensitivity 80.0%, specificity 92.3%). The AUC of 3 mRNA signature classifier is 0.904.
Discussion: Small amount changes of regulatory genes may have a greater impact on the biological effect. PCA have advantage in the preservation of original information, benefiting to preserve the effect to which regulatory genes contribute to AR, which partly contributed to the high accuracy of PCA-SVM classifier. HLA-DRB4, expressed in antigen presenting cells, presenting peptides of extracellular proteins, plays a crucial role in immune system, and associated with immune-related disease, such as type 1 diabetes and Anti-LGI1 encephalitis[2,3]. RPS4Y1, one of minor histocompatibility antigen genes, have shown strongly correlated with the kidney acute rejection in gender-mismatched transplantation[4]. The relationship between HLA-DRB4, SLC8A1 and kidney acute rejection has not been reported, and further research is needed
Conclusions: Supervised ML classifiers base on peripheral blood transcriptome data may be potential measures to monitor kidney allograft status early, accurately and dynamically.
Our gratitude goes to the GEO database provided comprehensive, multi-dimensional genomic data about kidney transplantation..
[1] [1] Johnson WE, Li C, Rabinovic A: Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007; 8: 118-127.
[2] [2] van Sonderen, A., et al., Anti-LGI1 encephalitis is strongly associated with HLA-DR7 and HLA-DRB4. Ann Neurol. 2017; 81(2): 193-198.
[3] [3] Zhao, L.P., et al., Eleven Amino Acids of HLA-DRB1 and Fifteen Amino Acids of HLA-DRB3, 4, and 5 Include Potentially Causal Residues Responsible for the Risk of Childhood Type 1 Diabetes. Diabetes. 2019; 68(8): 1692-1704
[4] [4] Tan, J.C., et al., H-Y antibody development associates with acute rejection in female patients with male kidney transplants. Transplantation. 2008; 86(1): 75-81